Stage 4 - Inversion and posterior uncertainty¶
Inversion¶
Once the HQ Calculations and Stage 3 - Observations, background and priors steps are completed, the final inversion step can be done. The user needs to also supply the values of the diagonal of R.
The program inversion.py performs the calculations, and makes several output files,
- shat.npy, shat.nc - The calculated
, both in numpy save format and netcdf format.
- hshat.txt -
convolved with H
- hqhti.npy - Result of
from Equation 1.
- hqht_zhsp.npy - Result of
from Equation 1.
- qhts.npy - The result of
from Equation 1.
Prerequisites¶
- R values - Variances of model data mismatch.
- sprior - contains sp. Can be either a numpy save file, dimensions (ntimesteps x ncells), or a netcdf file, with a variable named ‘sprior’ and dimensions (ntimesteps x nlat x nlon)
- HQ blocks created by hq.py.
- HQHT array created by hq.py.
- zhsp.txt created by make_z.py.
Usage¶
inversion.py is run with:
inversion.py [-c configfile] [-b] spriorfile rfile where Required arguments spriorfile : file name with sprior data rfile : file name with R data Optional arguments -c : Specify the configuration file to use. Default is 'config.ini'. -b : If set, use boundary value optimization files.
Posterior uncertainty¶
The uncertainty covariance of the estimated 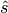 can be written as:
can be written as:
(1)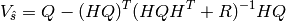
The a posteriori covariance matrix 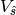 is a computational bottlenect for large inverse problems. Instead, the a posteriori
covariance is calculated directly at aggregated scales, without explicitly calculating
is a computational bottlenect for large inverse problems. Instead, the a posteriori
covariance is calculated directly at aggregated scales, without explicitly calculating 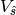 . The estimated aggregated grid-scale
. The estimated aggregated grid-scale
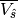 averaged over desired time periods is:
averaged over desired time periods is:
(2)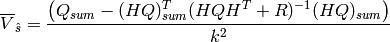
where 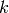 is the number of time steps in the aggregated time period. Using a space and time varying
is the number of time steps in the aggregated time period. Using a space and time varying 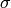 in equation 3,
in equation 3,
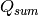 is defined as
is defined as
(3)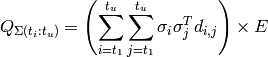
where:
(4)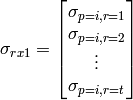
and 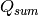 represents the sum of all Q blocks between
represents the sum of all Q blocks between 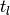 and
and 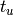 . However, greatly increased speed is achieved using the equivalent formulation:
. However, greatly increased speed is achieved using the equivalent formulation:
(5)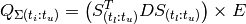
where 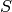 is a p✕r matrix such that:
is a p✕r matrix such that:
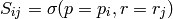
and 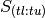 refers to the portion of S containing timesteps
refers to the portion of S containing timesteps 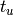 through
through 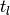 .
As in the constant
.
As in the constant 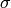 case,
case, 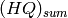 is computed simply by summing all of the column blocks of HQ between
is computed simply by summing all of the column blocks of HQ between 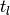 and
and 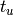 .
.
The time periods for calculating 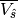 are defined in the configuration file with the key ‘vshat_dates’.
are defined in the configuration file with the key ‘vshat_dates’.
Example:
vshat_dates = ["2012-6-1", "2012-7-1", "2012-8-1", "2012-9-1"]
which will result in estimates for June, July and August of 2012. The time periods are from a date up to but not including the next date. Aggregated posterior uncertainties will also be computed for the entire time span defined in the configuration file with the keys ‘start_date’ and ‘end_date’.
The program apost.py performs the calculations. Output files created are
- vshat.npy, vshat.nc - Uncertainty estimates aggregated over the entire time of the inversion, in both npy and netcdf formats.
- vshat_month_1.npy, vshat_month_1.nc ... - Uncertainty estimates aggregated over each time period defined in the configuration file.
Usage¶
apost.py is run with:
apost.py [-c configfile] [-b] sigmafile where Required arguments sigmafile : file name with sprior sigma values Optional arguments -c configfile : Specify the configuration file to use. -b : If set, use boundary value optimization files.